Tech Support
Please contact Tong Hao with your questions or comments.
Please note: The web browser must be configured to accept cookies. And JavaScript must be enabled.
Newer versions:
hORFeome7.1
hORFeome8.1
Please note: The web browser must be configured to accept cookies. And JavaScript must be enabled.
Newer versions:
hORFeome8.1
Acknowledgments
The human ORFeome collection is made possible through support from the High-Tech Fund of the Dana-Farber Cancer Institute, The Ellison Foundation (Boston, MA), and by grants from the National Cancer Institute, the National Human Genome Research Institute, and the National Institute of General Medical Sciences.
About Human ORFeome
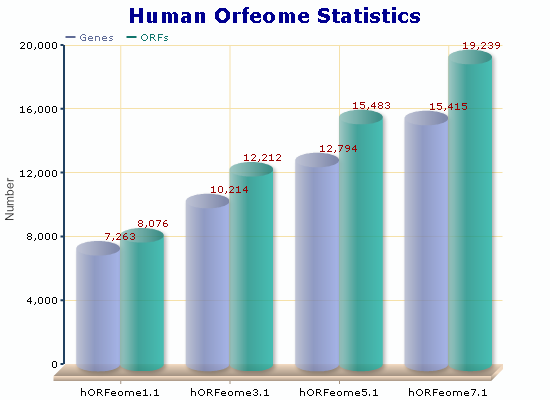
The advent of systems biology necessitates the cloning of nearly entire sets of protein-encoding open reading frames (ORFs) collected into ORFeome collections, so as to allow functional studies of the corresponding proteomes. Here we present a third version of the human ORFeome, human ORFeome 5.1.
With the Mammalian Gene Collection (MGC) resource as a starting point, we successfully cloned 15,483 human ORFs, representing at least 12,794 human genes, as mini-pools of PCR-amplified products. The human ORFeome version 5.1 (hORFeome v5.1) collection, includes the previous human ORFeome releases (v1.1 and 3.1), with the addition of 3,272 newly transferred ORFs from MGC templates.
hORFeome v5.1 represents a central resource of cloned human ORFs which can be transferred easily to Gateway compatible destination vectors for various functional proteomics studies. This set of ORFs ranges in size from 75 to more than 10Kb base pairs, and (excluding the RT-PCR rescued ORFs) contains 1,502 genes with multiple splice-variants and 814 polymorphic genes.
With the Mammalian Gene Collection (MGC) resource as a starting point, we successfully cloned 15,483 human ORFs, representing at least 12,794 human genes, as mini-pools of PCR-amplified products. The human ORFeome version 5.1 (hORFeome v5.1) collection, includes the previous human ORFeome releases (v1.1 and 3.1), with the addition of 3,272 newly transferred ORFs from MGC templates.
hORFeome v5.1 represents a central resource of cloned human ORFs which can be transferred easily to Gateway compatible destination vectors for various functional proteomics studies. This set of ORFs ranges in size from 75 to more than 10Kb base pairs, and (excluding the RT-PCR rescued ORFs) contains 1,502 genes with multiple splice-variants and 814 polymorphic genes.